
Our Research Lines

Artificial Intelligence in Health Care
Develop advanced analytical approaches and tools for improving diagnostics, treatment planning, and precision medicine.
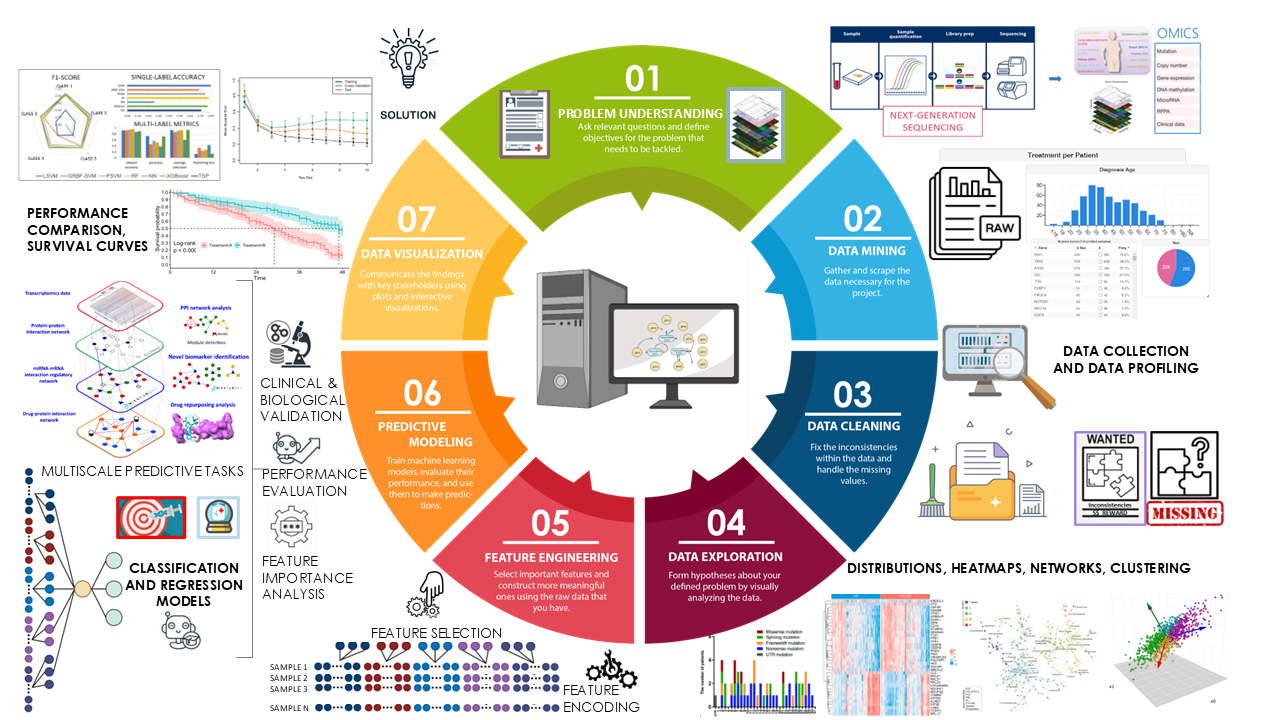
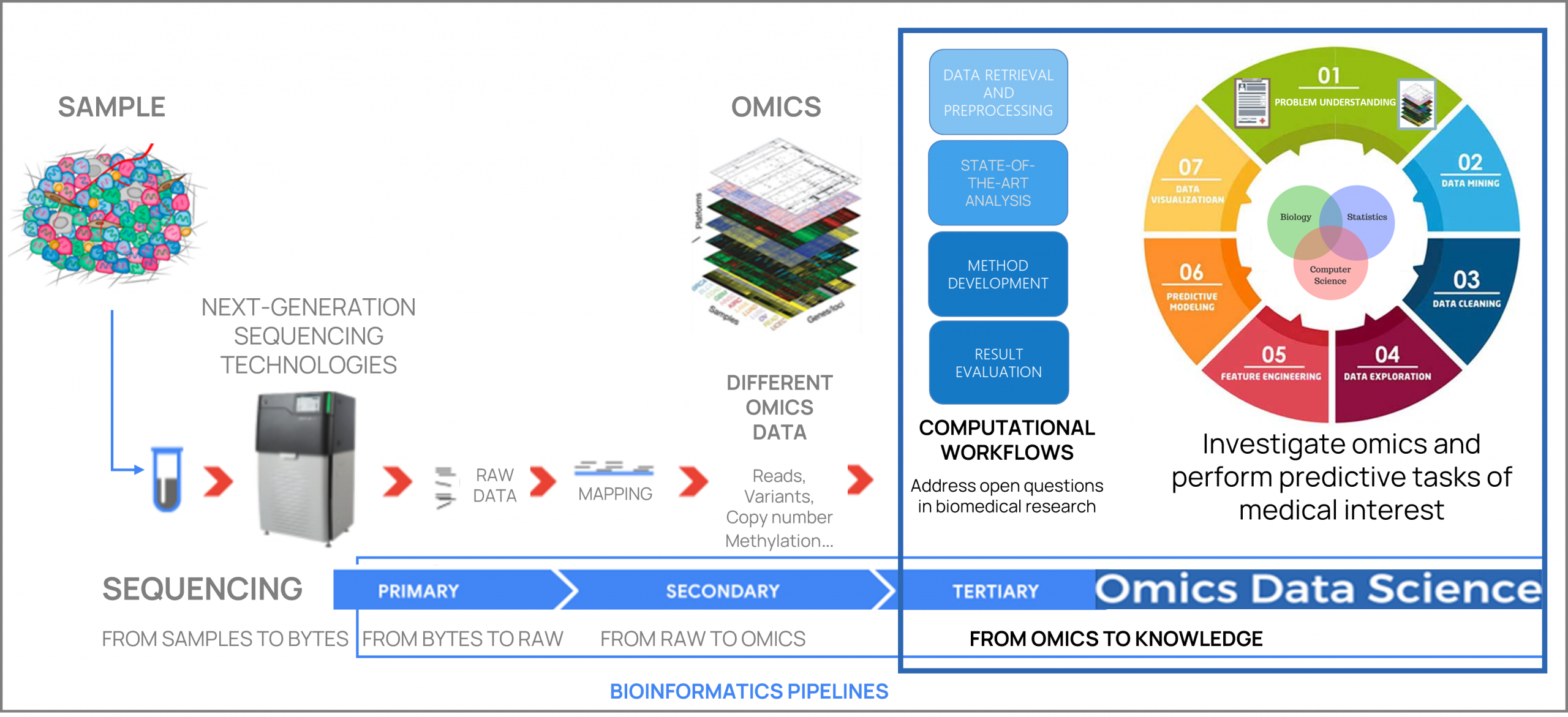
Omics Data Science
Leverage and design computational solutions to investigate large-scale omics data, and uncover insights to advance understanding of biological phenomena and diseases.

Computational Methods for Bioinformatics
Develop computational algorithms and tools to analyze heterogeneous data and interpret biological phenomena in bioinformatics.
Within these research lines, we integrate bioinformatics knowledge, mathematical and statistical data modelling, computer science principles, data analytics and machine learning approaches.
Topics actively investigated within our Research Lines
Main contributions:
- Machine learning for RNA sequencing-based intrinsic subtyping of breast cancer, Scientific Reports, 2020
- Investigating deep learning based breast cancer subtyping using pan-cancer and multi-omic data, IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2022
- Multi-label transcriptional classification of colorectal cancer reflects tumor cell population heterogeneity, Genome Medicine, 2023
- A novel machine learning-based workflow to capture intra-patient heterogeneity through transcriptional multi-label characterization and clinically relevant classification, Journal of Biomedical Informatics, 2025
Artificial Intelligence for Disease Subtyping & patient stratification
Within this research topic comprehensive omics-based studies and advanced computational methods are used to understand molecular intricacies underlying diseases, address biomedical issues and provide clinically relevant patient stratification contributing to precision medicine. In such a context, Machine Learning-based solutions emerged as a powerful key to dissect disease heterogeneity and unlock robust and clinically relevant patient predictions.
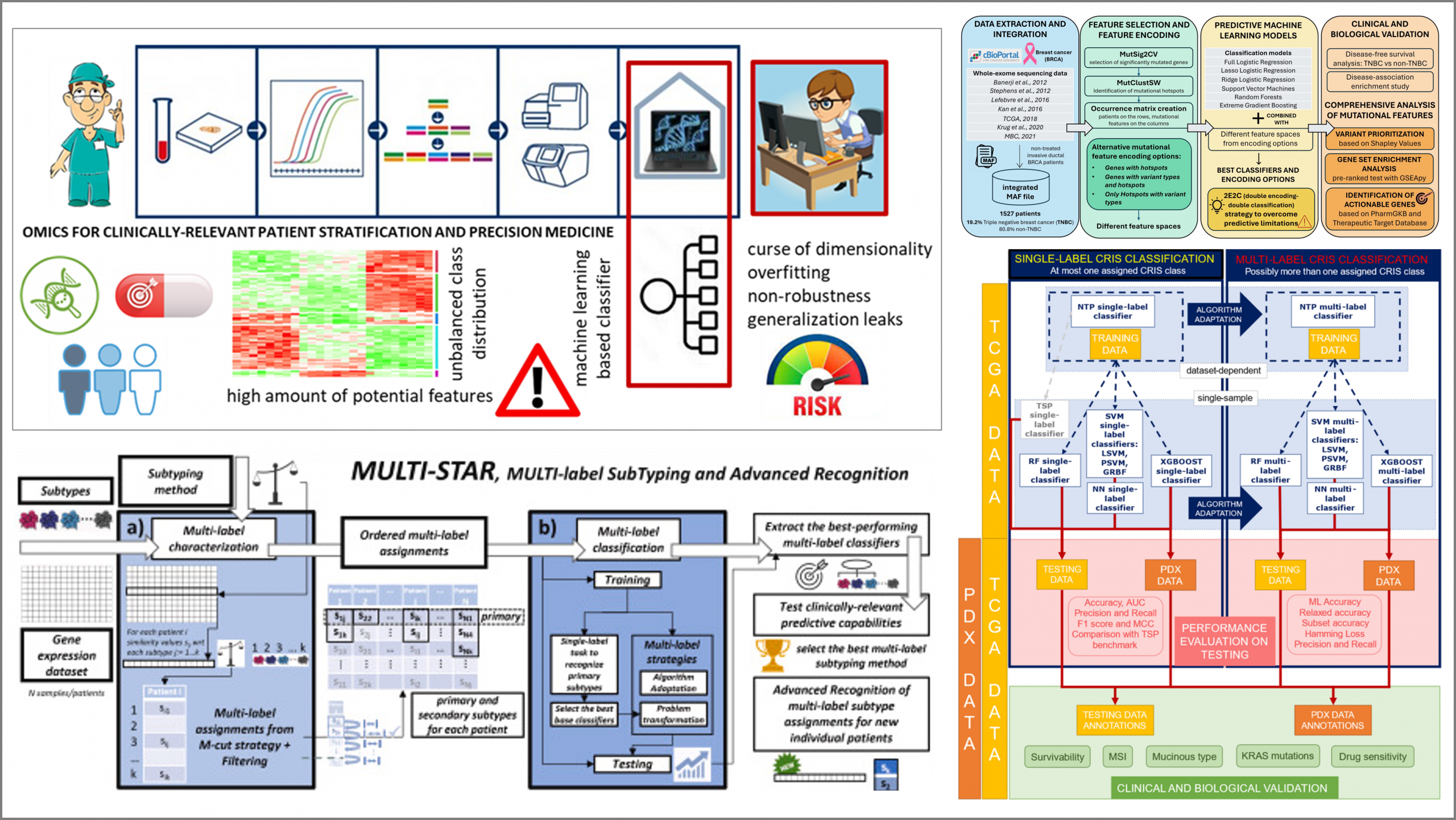
Main contributions:
- A data science approach to investigate the mutational landscape of a critical patient subgroup - Proceedings of the 19th International Conference on Computational Intelligence Methods for Bioinformatics and Biostatistics, CIBB 2024
- Three-stage data science methodology to explore genetic heterogeneity of diseases, Lecture Notes in Computer Science (LNBI) - Computational Intelligence Methods for Bioinformatics and Biostatistics, 2025
Omics Data Science for Tertiary analysis investigations
Within this research topic the typical Data Science life cycle is used to investigate hot topics in biomedical research. Attention is devoted to omics integration and exploration, feature engineering and predictive modelling. Result evaluation guarantees both computational and clinical-biological validation and explainability.
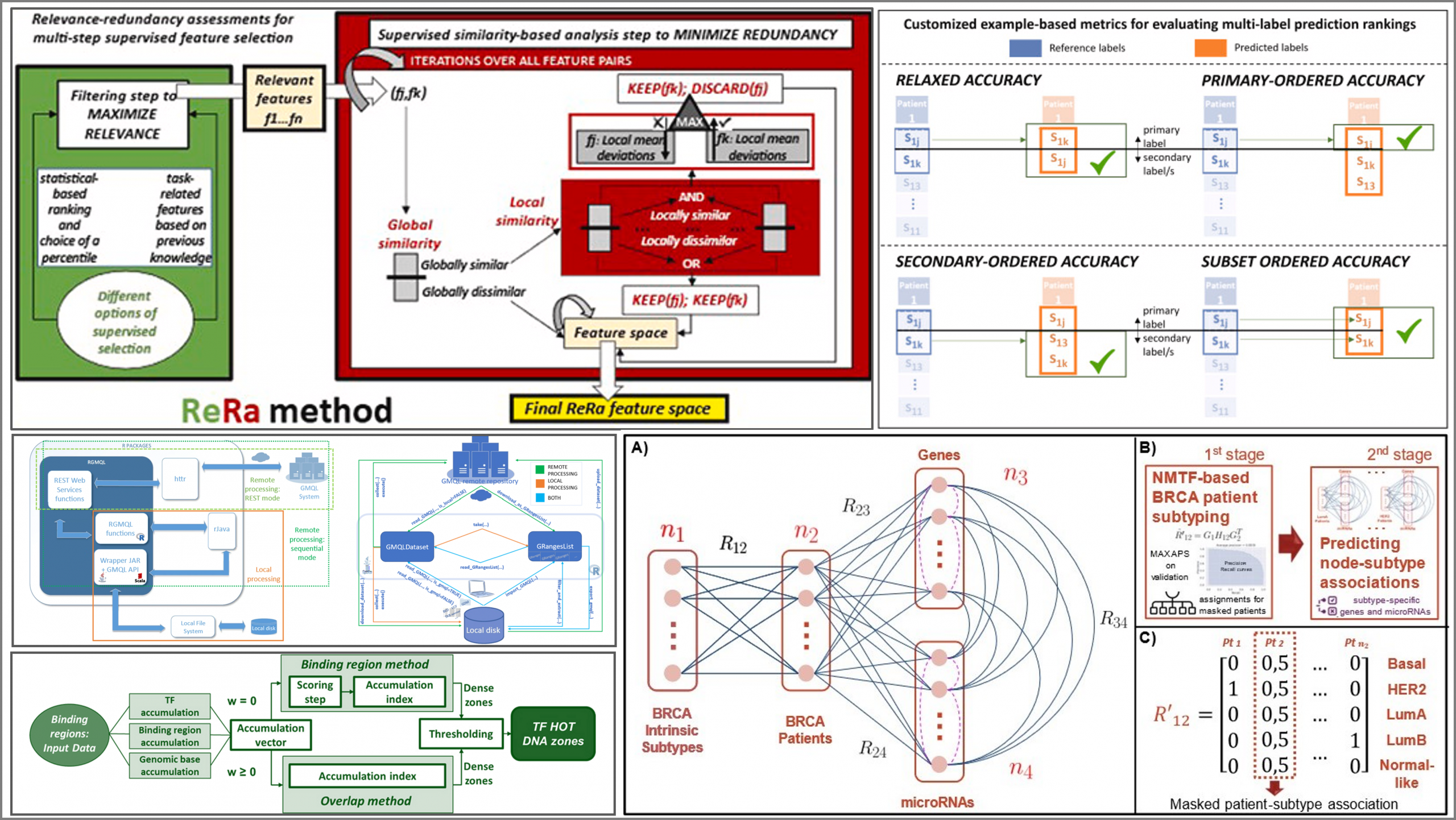
Main contributions:
- RGMQL: scalable and interoperable computing of heterogeneous omics big data and metadata in R/Bioconductor, BMC Bioinformatics, 2022
- Identification of transcription factor high accumulation DNA zones, BMC Bioinformatics, 2023
- Supervised Relevance-Redundancy assessments for feature selection in omics-based classification scenarios, Journal of Biomedical Informatics, 2023
- Inferring breast cancer subtype associations using an original omics integration based on Non-negative Matrix Tri-Factorization, Lecture Notes in Computer Science (LNBI) - Computational Intelligence Methods for Bioinformatics and Biostatistics, 2025
Computational Strategies for Bioinformatics & Health care
Within this research topic innovative computational strategies are designed and developed to answer unsolved questions or tackle specific challenges in Bioinformatics and Health Care domains. Achieving meaningful and reliable outcomes indeed requires leveraging advanced algorithms, scalable solutions, cutting-edge integration techniques, and rigorous analytical approaches that guarantee robustness, reproducibility, and interpretability across diverse and complex Bioinformatics & Health Care issues.
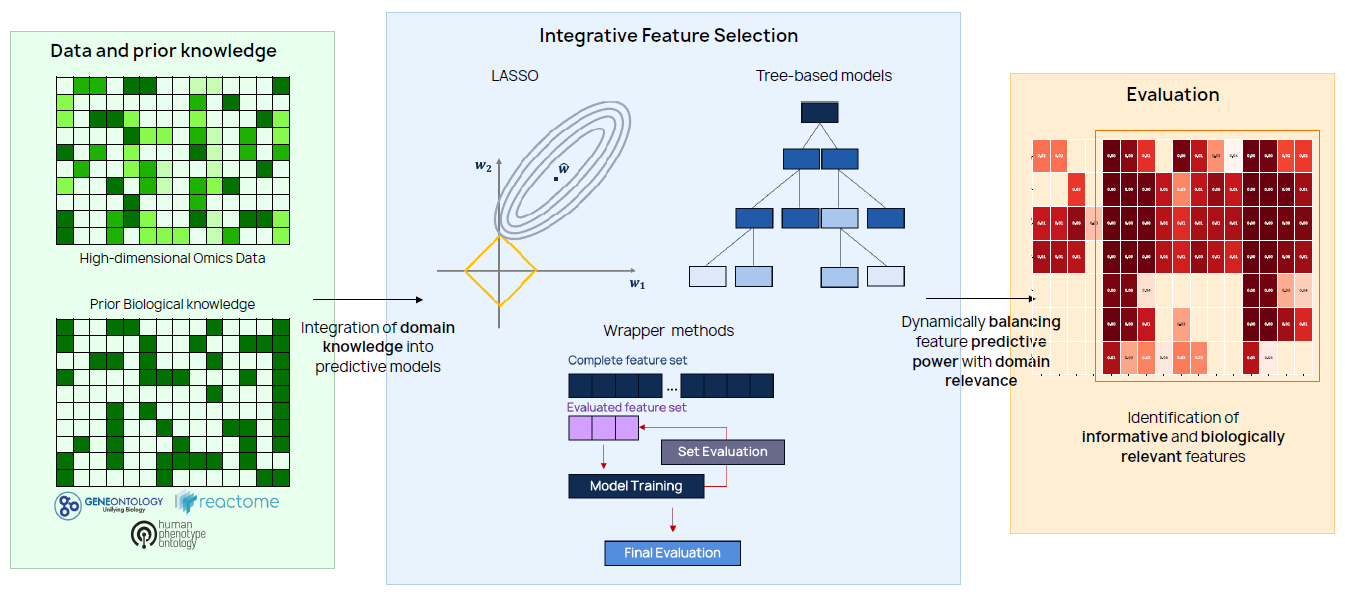
Main contributions:
- Biologically weighted LASSO: enhancing functional interpretability in gene expression data analysis, Bioinformatics, 2024
- Forward and backward feature selection guided by prior biological knowledge for enhanced interpretability, Lecture Notes in Computer Science (LNBI) - Computational Intelligence Methods for Bioinformatics and Biostatistics, 2025
- Improving tree-based models with biological insights: balancing predictivity and interpretability, Under review
Combining prior knowledge and feature selection
Within this research topic novel computational methods are developed to enhance the biological interpretability of predictive and feature selection algorithms in the
analysis of gene expression data leveraging prior biological knowledge.
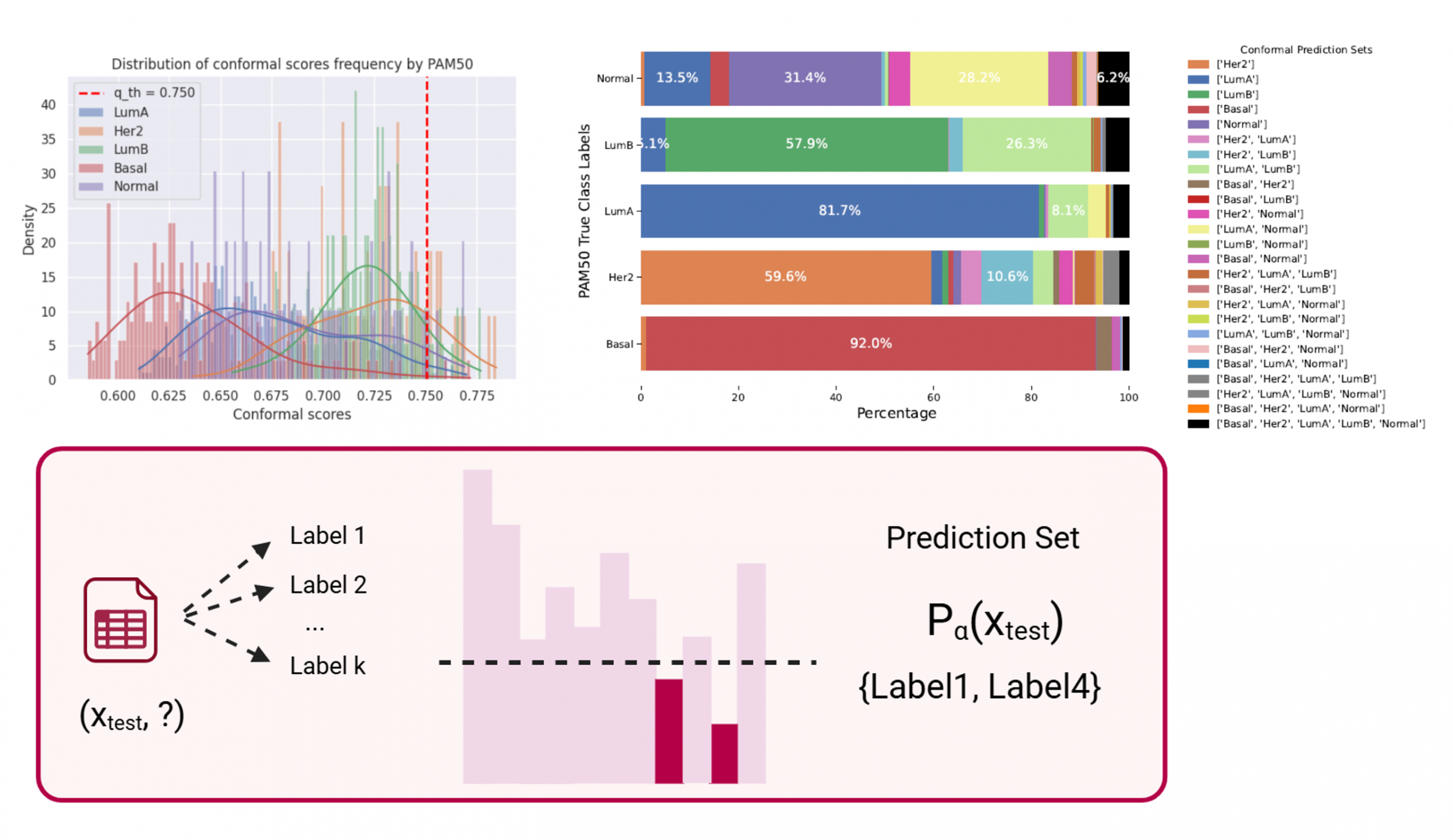
Example of conformal prediction in breast cancer subtype identification
Main contributions:
- Conformal prediction in breast cancer subtype identification - Proceedings of the 20th International Conference on Computational Intelligence Methods for Bioinformatics and Biostatistics, CIBB 2025
Conformal Prediction as a Tool for Precision Medicine
Within this research topic, conformal prediction is used to identify patient whose disease subtype classification may benefit from a multi-label approach rather than a single-label one. Outside its normal context of application, conformal prediction allows to highlight complex predictions on a patient-level.
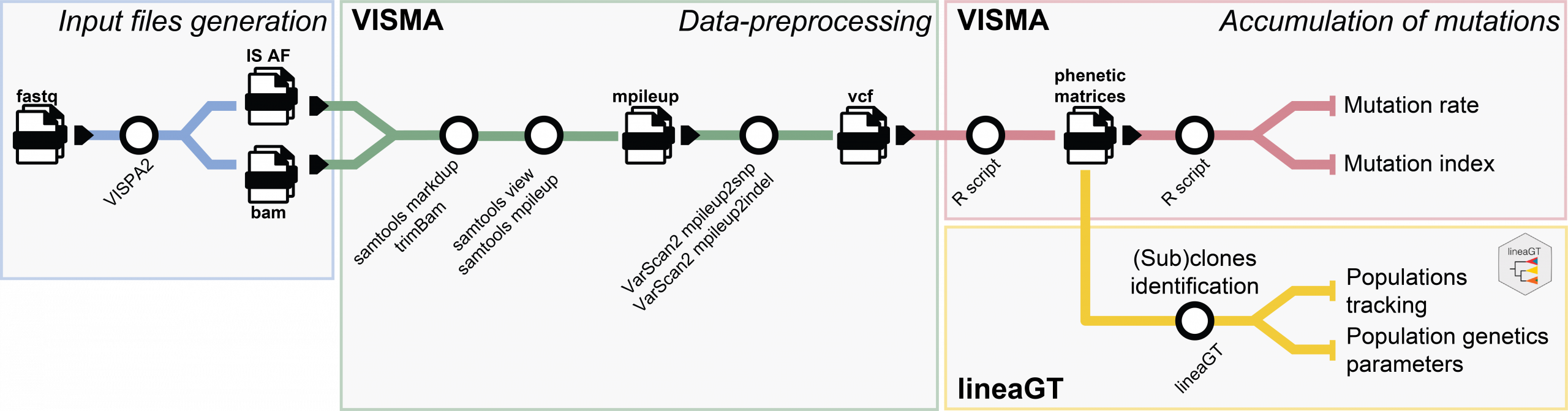
VISMA quantifies clone-resolved mutational burden over time, while lineaGT enables clonal and subclonal reconstruction to study hematopoietic dynamics in gene therapy and stem cell transplantation.
Main contributions:
- Vector insertional mutagenesis drives accelerated hematopoietic stem cell aging and acquisition of somatic mutations in vivo, Molecular Therapy, 28th Annual Meeting of the American Society of Gene and Cell Therapy. ASGCT, 2025.
- Deciphering hematopoiesis following ex-vivo gene therapy using Bayesian inference, Under review
- Insertional oncogene activation triggers non-cell-autonomous hematopoietic stem cell aging: Implications for genotoxicity beyond malignant transformation, Under review
Longitudinal clonal tracking methods using somatic mutations in patients under gene therapy and stem cell transplantation
Within this research topic, computational methods for longitudinal clonal tracking are developed by leveraging somatic mutations around lentiviral integration sites.
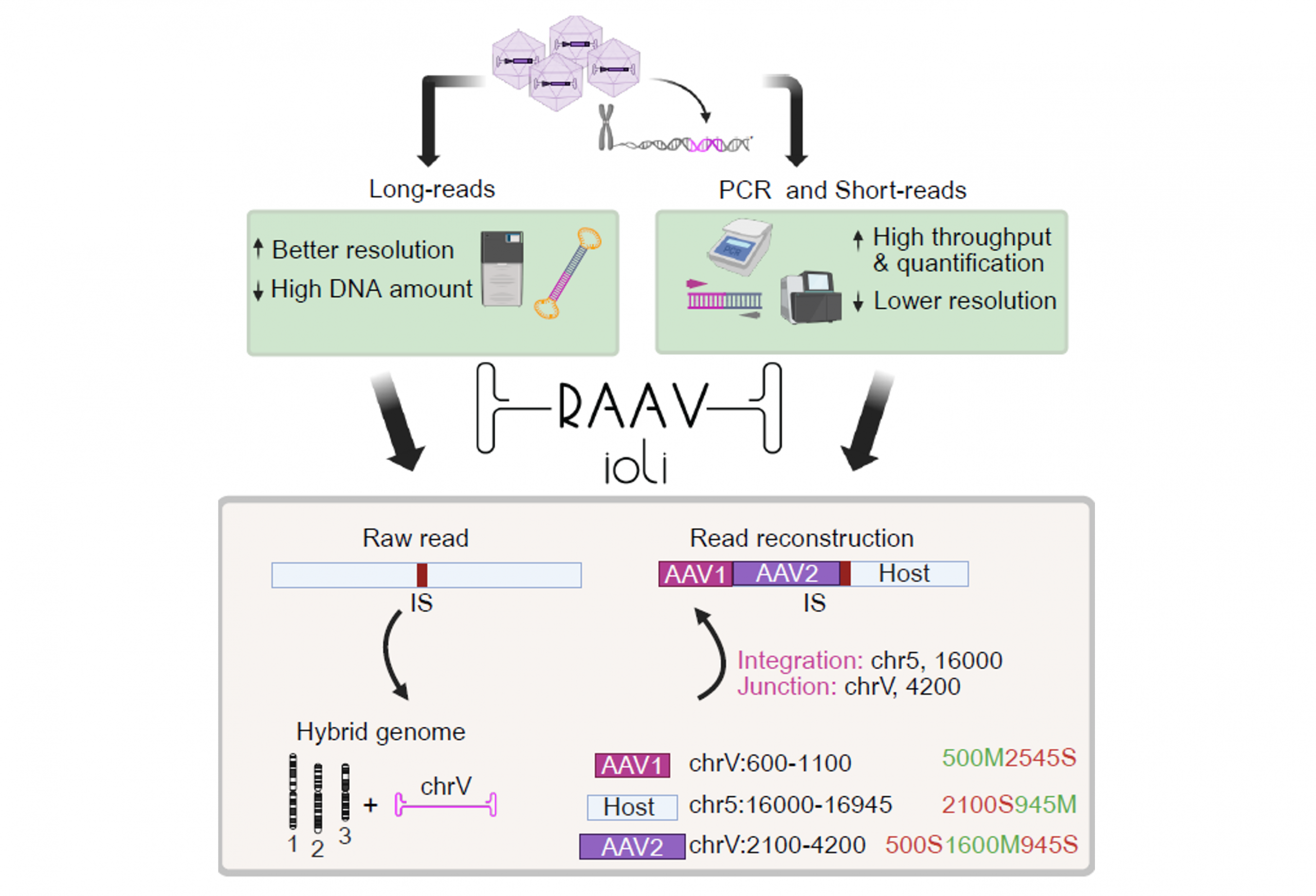
Main contributions:
- RAAVioli: A comprehensive approach to characterizing AAV vector integrations and rearrangements, Molecular Therapy Advances, 2026
- Profiling AAV integration in gene therapy and genome editing for Wilson disease, in Human Gene Therapy, ESGCT, 2025
comprehensive approaches to characterizing AAV vector integrations and rearrangements
Within this research topic, computational methods for AAV integration and rearrangement profiling are developed across sequencing platforms to detect AAV–genome junctions and reconstruct complex vector rearrangements enabling cross-context safety and efficacy monitoring.
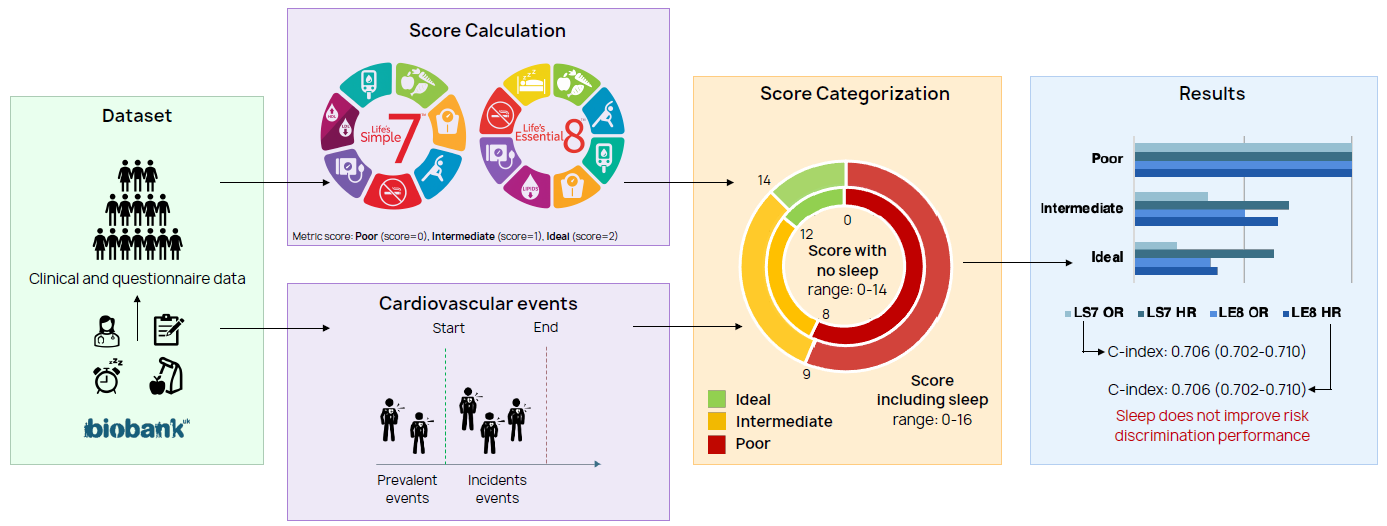
Main contributions:
- Validation of Life's Simple 7 and Life's Essential 8 in a European cohort: The role of sleep in cardiovascular risk estimation, Annual Congress of the European Association of Preventive Cardiology (EAPC), ESC Preventive Cardiology 2025
- Understanding the impact of sleep on cardiovascular risk estimation: comparison of LS7 and LE8 performances in a European population, Under review
Cardiovascular risk prediction
Within this research topic computational models for cardiovascular risk prediction are evaluated to beter identify patients at high risk of developing fatal and non-fatal cardiovascular disease (CVD) events, with a particular focus on incorporating additional potential risk factors.
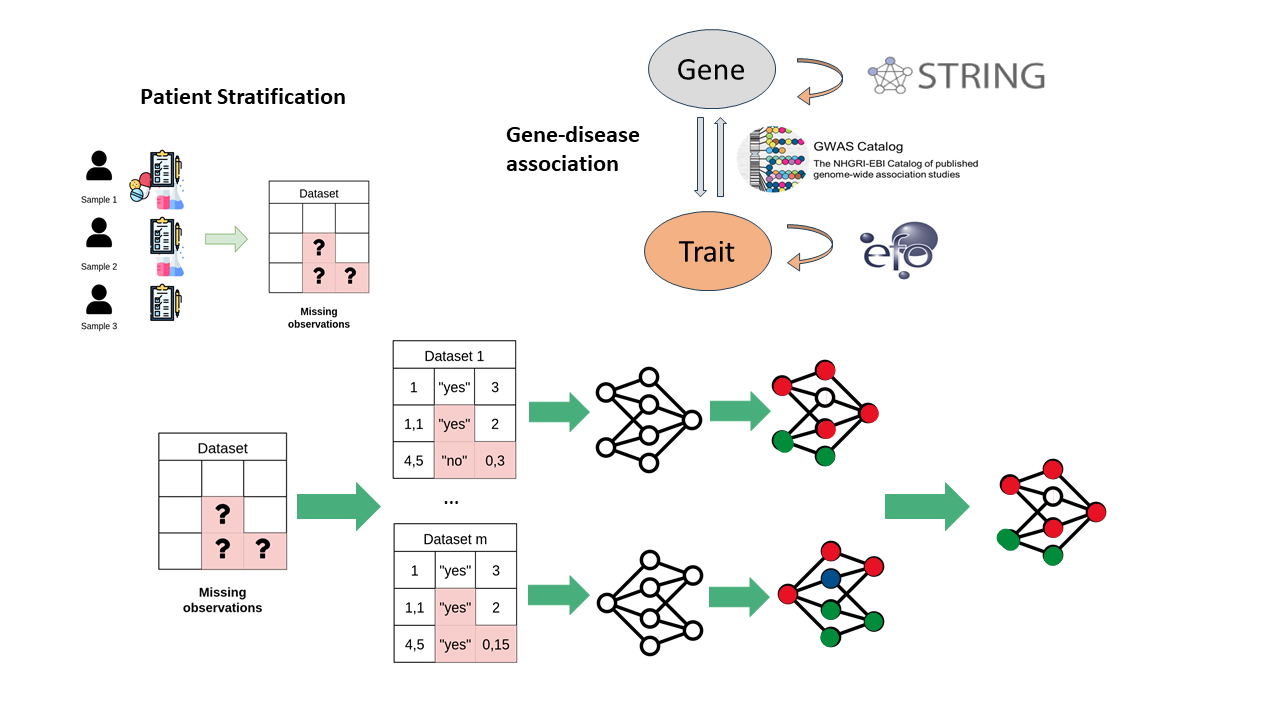
Examples of studied pathologies include Chronic Pain, Myotonia, and Amyotrophic Lateral Sclerosis.
Integrative Data Approaches for Neurological Disorder Insights
Within this research topic, various supervised and unsupervised techniques are explored and adapted to uncover the molecular mechanisms underlying pathological disorders. Special emphasis is placed on integrative approaches that model complex system interactions as networks and leverage this complexity through computational methods.
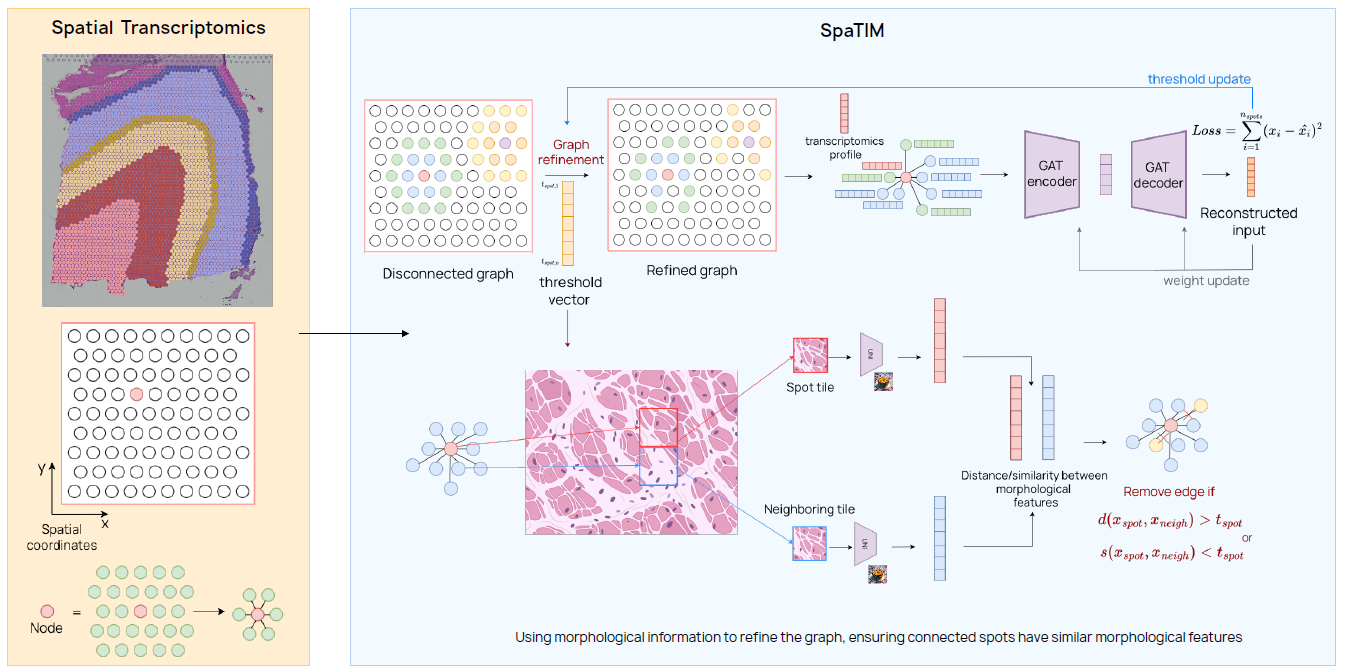
Spatial transcriptomics analysis
Within this research topic novel computational approaches for the analysis of spatial transcriptomics (ST) data are developed leveraging Graph Neural Networks (GNNs) and
incorporating additional biological and morphological context to improve graph construction and representation learning.
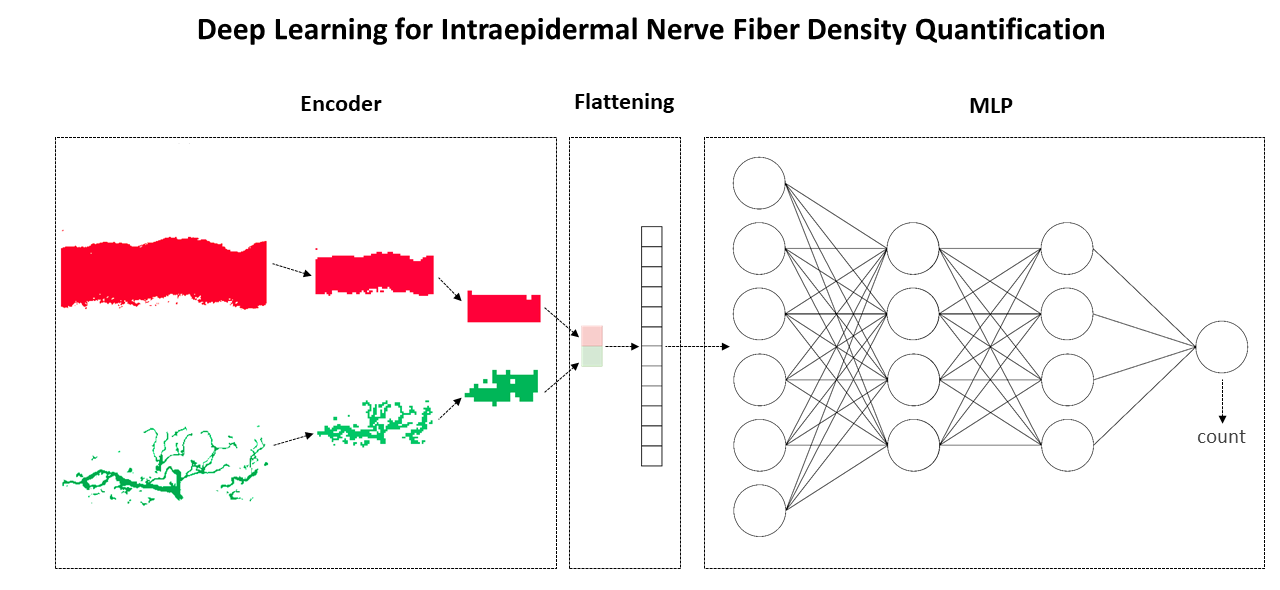
Applications include deep learning–based classification of alpha-synucleinopathies and assessment of intraepidermal nerve fiber density, useful for small-fiber neuropathy evaluation.
Deep Learning–Driven Analysis of Skin Biopsies
Within this research topic, modern deep learning techniques are applied to confocal images of skin biopsies. Evaluating skin biopsies for diagnostic purposes offers a low-cost alternative to Magnetic Resonance Imaging, though it remains operator-dependent and not widely standardized.
